Interactome INSIDER
About Downloads Home
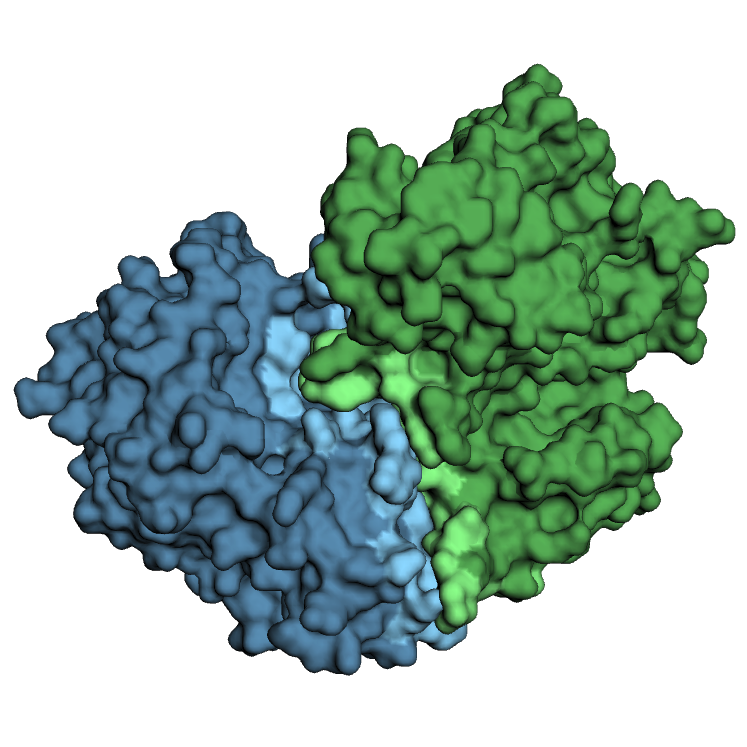
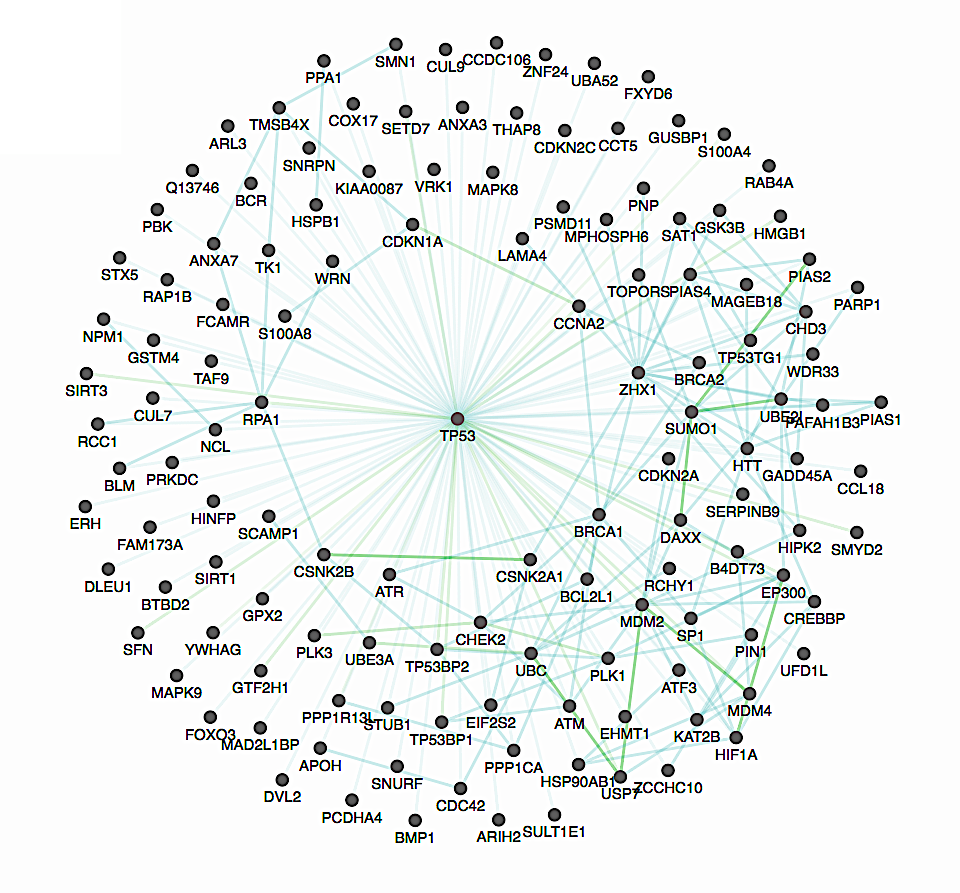
Co-crystal structures are the most reliable source of partner-specific interface residue information. We compile all available co-crystal structures from the RCSB Protein Data Bank, resolving the interface of 7,135 interactions through their 3D structures.
We compile a collection of more than 56,000 disease-associated mutations from both HGMD and ClinVar, additionally processing more than 1.3 million somatic mutations from COSMIC. These mutations can be explored in the context of interface domains, residues, or 3D atomic clustering, providing multi-scale information for genomic studies.
We compile more than 600,000 Population variants from all populations contained in the 1000 Genomes Project, ESP, and UniProt. Users can explore selective pressure in the context of both interface residues and disease mutations.